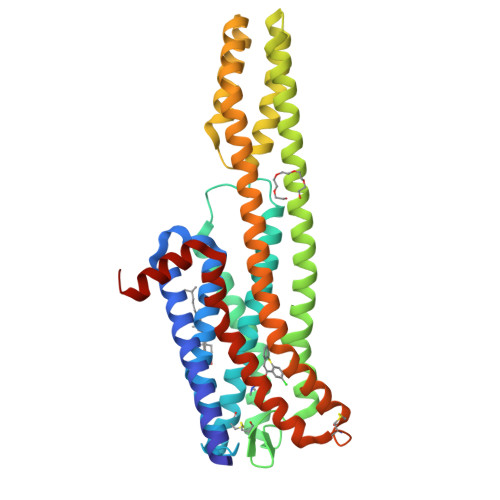
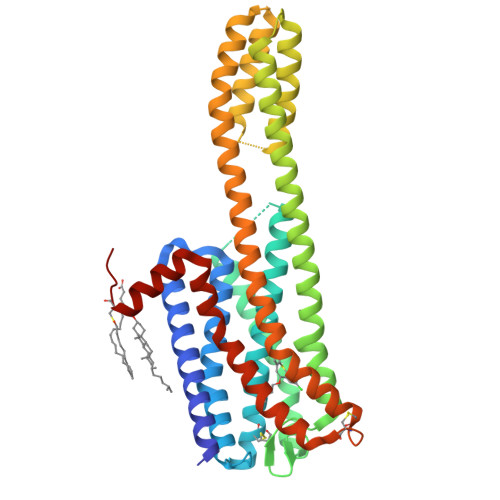
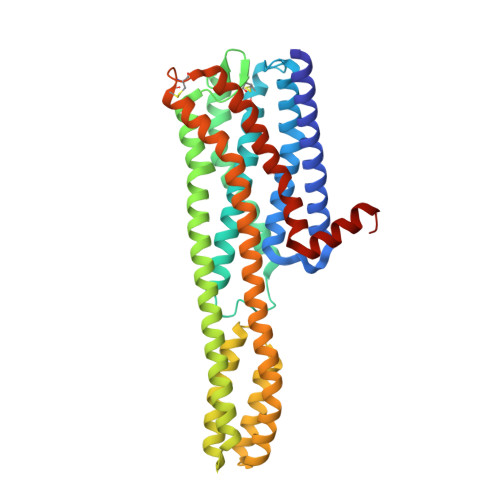
Structures of the 5-HT2Areceptor in complex with the antipsychotics risperidone and zotepine.
Kimura, K.T., Asada, H., Inoue, A., Kadji, F.M.N., Im, D., Mori, C., Arakawa, T., Hirata, K., Nomura, Y., Nomura, N., Aoki, J., Iwata, S., Shimamura, T.(2019) Nat Struct Mol Biol 26: 121-128
- PubMed: 30723326
- DOI: https://doi.org/10.1038/s41594-018-0180-z
- Primary Citation of Related Structures:
6A93, 6A94 - PubMed Abstract:
Many drugs target the serotonin 2A receptor (5-HT 2A R), including second-generation antipsychotics that also target the dopamine D 2 receptor (D 2 R). These drugs often produce severe side effects due to non-selective binding to other aminergic receptors. Here, we report the structures of human 5-HT 2A R in complex with the second-generation antipsychotics risperidone and zotepine. These antipsychotics effectively stabilize the inactive conformation by forming direct contacts with the residues at the bottom of the ligand-binding pocket, the movements of which are important for receptor activation. 5-HT 2A R is structurally similar to 5-HT 2C R but possesses a unique side-extended cavity near the orthosteric binding site. A docking study and mutagenic studies suggest that a highly 5-HT 2A R-selective antagonist binds the side-extended cavity. The conformation of the ligand-binding pocket in 5-HT 2A R significantly differs around extracellular loops 1 and 2 from that in D 2 R. These findings are beneficial for the rational design of safer antipsychotics and 5-HT 2A R-selective drugs.
Organizational Affiliation:
Department of Cell Biology, Graduate School of Medicine, Kyoto University, Kyoto, Kyoto, Japan.